MaizegoSummer2022
2. Homology-based annotation
-
Evidences (Proteins) collection and remove redundants
-
Proteins align to genome
-
Parse alignment and generate annotate regions
- or based on WGA with well-annotated genomes (not commonly used)
Common Softwares:
- Dedup:
CD-HIT - Mapping:
Diamond/Blastp,Exonerate... - WGA:
blat,last,minimap2,mummer,anchorwave...
MaizegoSummer2022
3. EST evidences (RNA-Seq)
- Collecting public data (ESTs)
- RNA-Sequencing: 2nd-generation, 3rd-generation
- De novo assembly, Genome-guide assembly
- **Transcript-mapping" against genome
- Parse alignment
- Refining
Common Softwares:
- RNA-seq mapping:
minimap2,hisat2,star... - Assembly:
stringtie,Cufflinks... - Mapping transcripts:
GMAP,minimap2... - Pipelines:
PASA
MaizegoSummer2022
4. Combine all evidences
Evidence confidence level
- ISO-seq (3rd generation RNA): high-quality, isoforms, UTRs; BUT rare;
- 2nd generation RNA-seq: Some UTRs, reliable structures, some isoforms; Common;
- Homolog evidences
- Predictions: Poor on isoforms
Common Softwares:
- Combine Evidences:
EVM,MAKER...
MaizegoSummer2022
Before gene annotation:
1. Genome Assembly Estimation
- Low quality genomes lead to wrong gene annotation
2. TE annotation and repeatmasking
- Another important genome annotaton area we will learn in the future
- Mask to reduce calculating time, and noises
- Do not mask simple and low repeats (some genes may contain these structure)
MaizegoSummer2022
Along-side or After gene annotation
Along-side:
-
tRNA and ncRNA annotation
tRNA: tRNA-scanncRNA: Rfam HMM models
After:
Filtering: low-evidence models, TE-overlap models, pre-mature models ...- Functional annotation and downstream analyses ...
MaizegoSummer2022
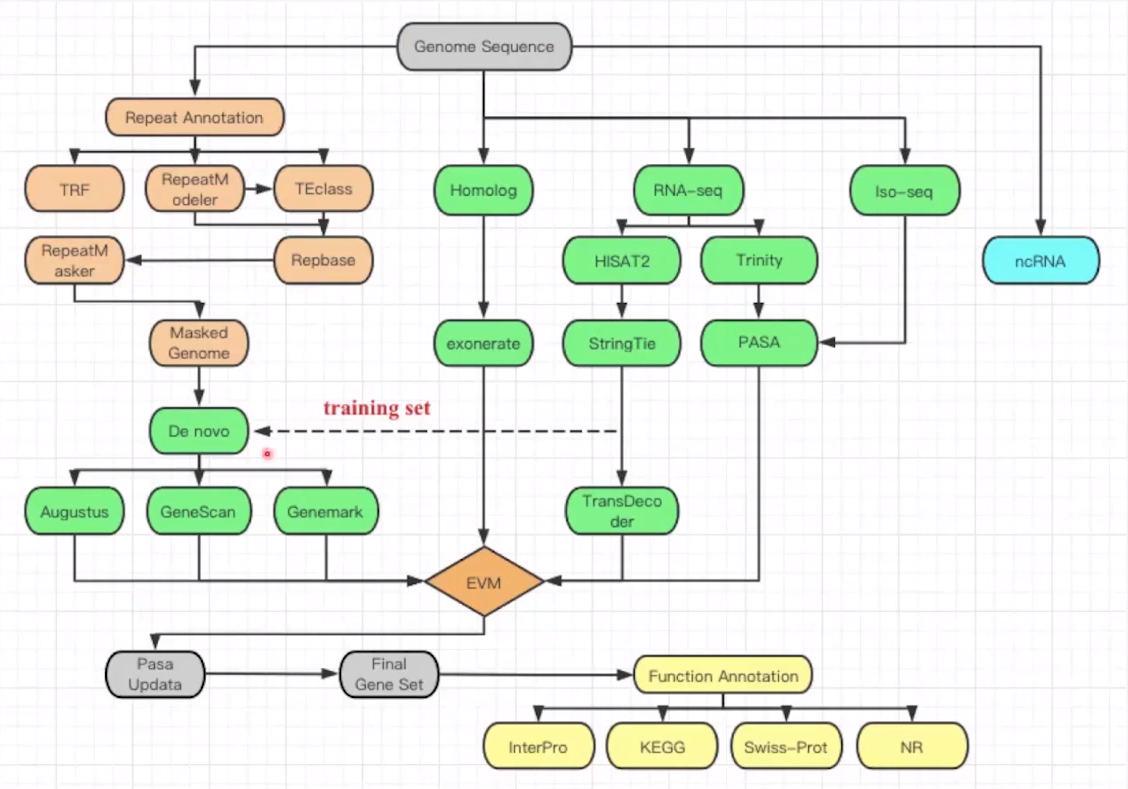
Overview of gene annotation pipelines:
MaizegoSummer2022
Common wheels
-
MAKER: maybe the most used annotation tool, tutorials on NCPGR WIKI -
Funannotate: newly friendly pipeline wrapper Github - ...
MaizegoSummer2022